ANNEX V(Article 5)
DETERMINATION OF THE CONTENT OF ENANTHIC ACID TRIGLYCERIDE IN BUTTER, BUTTER-OIL AND CREAM BY GAS CHROMATOGRAPHIC ANALYSIS OF TRIGLYCERIDES
1.SCOPE
This method lays down a method for the determination of the content of the triglyceride of enanthic acid in butter-oil, butter and cream.
2.TERMS AND DEFINITION
Enanthic acid content: content of the triglyceride of enanthic acid determined by the procedure specified in this method.
Note: The enanthic acid content is expressed in kg per ton of product for butter-oil and butter, and it is expressed in kg per ton of milk fat for cream.
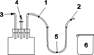
3.PRINCIPLE
Milk fat is extracted from the different products according to ISO 14156 | IDF 172:2001. The quantitative determination of the content of the triglyceride of enanthic acid in the extracted fat is determined by capillary gas chromatography (GC). The result obtained for the sample is evaluated by reference to the triglyceride of caproic acid as internal standard.
Note: Tributyrin has also been found to be a satisfactory internal standard.
4.REAGENTS
Use only reagents of recognized analytical grade.
4.1.n-Hexane
4.2.Standard triglyceride of caproic acid, at least 99 % pure
4.3.Standard triglyceride of enanthic acid, at least 99 % pure
4.4.Anhydrous sodium sulfate (Na2SO4).
5.APPARATUS
Usual laboratory equipment and particularly the following:
5.1.Analytical balance precise at 1 mg
5.2.Volumetric flasks, of capacities 10 ml and 20 ml
5.3.Tubes for centrifuge, of capacity 30 ml
5.4.Rotary evaporator
5.5.Oven, capable of being maintained at a temperature of 50 °C ± 5 °C
5.6.Filter paper, medium porosity, of diameter about 15 cm
5.7.
Gas chromatography equipment
5.7.1.Gas chromatograph equipped with a split/splitless or on-column injector and a flame ionization detector (FID)
5.7.2.
GC column, with a stationary phase which has successfully employed to perform triglyceride separation (100 % dimethylpolysiloxane or 5 % phenyl-95 % methylpolysiloxane). Select the stationary phase, the column length (between 4 m and 15 m), the internal diameter (between 0,22 mm and 0,50 mm) and the film thickness (0,12 μm or more) taking into account the laboratory experience and the injection system applied. In any case the selected column shall produce both a complete separation between the solvent peak and the triglyceride of caproic acid and a baseline resolution between triglyceride of caproic and enanthic acid peaks. Examples of applicable conditions are listed below.
5.7.2.1.Example of applicable conditions using a split injector:
Carrier gas: helium
Column head pressure: 100 KPa
Column: 12 m length, 0,5 mm internal diameter, 0,1 μm film thickness fused silica column
Stationary phase: 100 % dimethylpolysiloxane or 5 % phenyl-95 % dimethylpolysiloxane (for ex. HT5)
Column temperature: initial temperature of 130 °C, maintained for 1 min, raised at a rate of 20 °C/min up to 260 °C and then raised at a rate of 30 °C/min up to 360 °C; maintain 10 mn at 360 °C
Detector temperature: 370 °C
Injector temperature: 350 °C
Split ratio 1:30
Amount of sample injected: 1 μl.
5.7.2.2.Example of applicable conditions using an on-column injector:
Carrier gas: hydrogen (constant flow system)
Column head pressure: 89 kPa
Column: 4 m length, 0,32 mm internal diameter, 0,25 μm film thickness, fused silica column
Stationary phase: 5 % phenyl, 95 % dimethylpolysiloxane
Column temperature: initial temperature of 60 °C, maintained for 2 min, raised at a rate of 35 °C/min up to 340 °C, maintained at this temperature for 5 min
Detector temperature: 350 °C
Amount of sample injected: 1 μl
5.8.Injection syringe, of capacity 5 μl.
6.SAMPLING
It is important that the laboratory receives a sample which is truly representative and has not been damaged or changed during transport or storage.
Sampling is not part of the method specified in this International Standard. A recommended sampling method is given in IDF: standard 50C:1995 or ISO 707-1997 — Milk and milk products — Methods of sampling.
7.PROCEDURE
7.1.Preparation of the test sample and test portion
Proceed according to ISO 14156 | IDF 172:2001
7.1.1.Butter-oil, Butter
7.1.1.1.Melt 50 g to 100 g of test sample in the oven (5.5)
7.1.1.2.Place 0,5 g to 1,0 g of anhydrous sodium sulfate (5.4) in a folded filter paper
7.1.1.3.Filter the fat through the filter paper containing anhydrous sodium sulfate collecting the filtrate in a beaker maintained in the oven (5.5). When decanting the melted butter onto the filter paper, take care that no serum is transferred
7.1.2.Cream
7.1.2.1.Bring the test sample to a temperature of 20 °C ± 2 °C
7.1.2.2.Mix or stir the sample thoroughly
7.1.2.3.Dilute a suitable amount of test sample so as to obtain 100 ml of test portion with a mass fraction of fat of approximately 4 %
7.1.2.4.Proceed as with raw milk and homogenized milk (see ISO 14156 | IDF 172:2001, §8.3) to extract the fat from the cream
7.1.2.5.Weigh in a 10 ml volumetric flask (5.2), to the nearest 1 mg, 1 g of the extracted fat. Add 1 ml of the solution 7.2.2. Complete to 10 ml with n-hexane (4.1) and homogenise
7.1.2.6.Introduce 1 ml of the solution 7.1.1.2 in a 10 ml volumetric flask (5.2) and dilute to 10 ml with n-hexane (4.1)
7.2.Preparation of the calibration standards
7.2.1.Dissolve 100 mg of the triglyceride of enanthic acid (4.3) in 10 ml of n-hexane (4.1)
7.2.2.Dissolve 100 mg of the triglyceride of caproic acid (4.2) in 10 ml of n-hexane (4.1)
7.2.3.Introduce 1 ml of the solution 7.2.2 in a 10 ml volumetric flask (5.2). Complete to 10ml with n-hexane (4.1)
7.2.4.Introduce 1 ml of the solution 7.2.1 and 1 ml of the solution 7.2.2 in a 10 ml volumetric flask (5.2). Complete to 10 ml with n-hexane (4.1)
7.2.5.Introduce 1 ml of the solution 7.2.4 in a 10 ml volumetric flask (5.2) and complete to 10 ml with n-hexane (4.1)
7.3.Chromatographic determination
7.3.1.Inject 1 μl of the standard solution 7.2.5 twice
7.3.2.Inject 1 μl of each sample solution
Note: If the on column injector system is adopted an increased dilution should be applied both to the standard and sample solutions.
7.3.3.Repeat the operation 7.3.1 every 3 samples in order to bracket samples between duplicate standard injections. Results are based upon the mean average response factors from the standard chromatograms.
8.CALCULATION OF RESULTS
For each chromatogram, integrate the area of the peaks associated with the triglycerides of enanthic acid and caproic acid.
Follow those instructions for each bracketed sequence i.e. for a set of bracketed samples, the standard injected twice immediately before them is STD1 and the standard injected twice immediately after them is STD2.
8.1.Calibration
8.1.1.Calculate the response factor for each duplicate of STD1, Rf1(a) and Rf1(b)
Rf1 (a) or (b) = (Peak area for caproic acid triglyceride/Peak area for enanthic acid triglyceride) × 100
Calculate the mean average response factor, Rf1
Rf1 = (Rf1(a) + Rf1(b)) / 2
8.1.2.Similarly, calculate the mean average response factor STD2, Rf2
8.1.3.Calculate the mean average response factor, Rf
Rf = (Rf1 + Rf2) /2
8.2.Test samples
For each sample chromatogram obtained between STD1 and STD2, calculate the enanthic acid content, C (kg/t):
C = (Peak area for enanthic acid triglyceride × Rf × 100)/(Peak area for caproic acid triglyceride × Wt × 1 000)
where:
Wt = weight of fat taken (g),
100 = dilution volume for sample,
1 000 = conversion factor (for μg/g to kg/t)
For butter samples, take the fat content of butter into account and calculate a corrected concentration value, Cbutter (kg/t of butter)
Cbutter = Cfat × F
where F is the fat content of butter.
9.PRECISION
Details of an interlaboratory test on butter in accordance with ISO 5725-1 and ISO 5725-2 on the precision method are shown in (12.).
The values for repeatability and reproducibility limit are expressed for the 95 % probability level and may not be applicable to concentration ranges and matrices other than those given.
9.1.Repeatability
The absolute differences between two individual single test results, obtained with the same method on identical test material in the same laboratory by the same operator using the same equipment within a short interval of time, will in not more than 5 % of cases be greater than 0,35 kg/t.
9.2.Reproducibility
The absolute differences between two individual single test results, obtained with the same method on identical test material in different laboratories with different operators using different equipment will in not more than 5 % of cases be greater than 0,66 kg/t.
10.TOLERANCE LIMITS: LOWER LIMITS (CASE OF INSUFFICIENT QUANTITIES)
10.1. Three samples must be taken from the traced product in order to check on the correct tracing of the product
10.2.Butter and concentrated butter
10.2.1.The incorporation rate is 11 kg of at least 95 % pure enanthic acid triglyceride per tonne of butter, i.e. 10,45 kg/t
10.2.2.The results of three samples obtained from the analysis of the product are used to check the rate and the homogeneity of tracer incorporation and the lowest of these results is compared with the following limits:
9,51 kg/t (95 % of the minimum incorporation rate of 95 % pure enanthic acid triglyceride, single determination),
6,89 kg/t (70 % of the minimum incorporation rate of 95 % pure enanthic acid triglyceride, single determination),
The tracer concentration of the sample giving the lowest result is used in conjunction with interpolation respectively between 9,51 kg/t and 6,89 kg/t.
10.3.Cream
10.3.1.The incorporation is 10 kg of at least 95 % pure enanthic acid triglyceride per tonne of milk fat, i.e. 9,50 kg/t traced milk fat
10.3.2.The results of the three samples obtained from the analysis of the product are used to check the rate of the homogeneity of tracer incorporation and the lowest of these results is compared with the following limits:
8,60 kg/t (95 % of the minimum incorporation rate of 95 % pure enanthic acid triglyceride, single determination),
6,23 kg/t (70 % of the minimum incorporation rate of 95 % pure enanthic acid triglyceride, single determination),
The tracer concentration of the sample giving the lowest result is used in conjunction with interpolation respectively between 8,60 kg/t and 6,23 kg/t.
11.TOLERANCE LIMITS: UPPER LIMITS (CASE OF EXCEEDING QUANTITY BY MORE THAN 20 %)
11.1. Three samples must be taken from the traced product in order to check on the correct tracing of the product
11.2.Butter and concentrated butter
11.2.1.The results of three samples obtained from the analysis of the product are used to check the rate and the homogeneity of tracer incorporation and the mean of these results is compared with the following limits:
Upper limit is 12,96 kg/t
11.3.Cream
11.3.1.The results of three samples obtained from the analysis of the product are used to check the rate and the homogeneity of tracer incorporation and the mean of these results is compared with the following limits:
Upper limit is 11,82 kg/t.
12.ADDITIONAL INFORMATION: STATISTICAL ANALYSIS OF RESULTS ON THE DETERMINATION OF TRIENANTOATE IN BUTTERFAT BY TRIGLYCERIDE ANALYSIS
Four collaborative trials have been carried out to determine the trienantoate content in traced butter.
Nine laboratories participated to the 1st ring test and no specifications were provided about the analytical methods to use:
10 laboratories participated to the 2nd ring test and 4 different methods were applied:
Quantification of methylheptanoate by using n-nonane or methylnonanoate as internal standard
Quantification of trienantoate by using tricaproate as internal standard
Quantification of methylheptanoate by using a calibration sample/mixture
Quantification of methylheptanoate by using a calibration mixture.
Moreover, if FAME were analysed, two different methylation procedures were used (De Francesco and Christopherson & Glass).
Due to the results obtained, two methods were chosen to perform the 3rd ring test:
Quantification of methylheptanoate by using n-nonane or methylnonanoate as internal standard
Quantification of trienantoate by using tricaproate as internal standard.
The results of 7 labs showed that the FAME method produced a higher variability and consequently it was decided to use only the determination of trienantoate as triglyceride following the procedure of the q Quantification of trienantoate by using tricaproate as internal standard. Moreover the triglyceride analysis has to be carried out by capillary column.
In the 4th ring test four samples (A, B, C, D) were circulated and nine laboratories provided results (Tables 1-2).
Two laboratories (DE and UE) analysed the samples by using FAME method.
Due to the reduced number of laboratories, the Statistical calculation has been performed both on the complete set (Figures 1-2) of data including FAME results and on the data obtained from TG analysis.
Tests for outliers:
sample A. Dixon, Cochran and Grubbs tests at levels 1 and 5 %, showed one laboratory outlier.
sample B. Grubbs test at level 5 % showed one laboratory outlier.
sample C. Dixon and Grubbs tests at levels 1 and 5 %, showed one laboratory outlier.
sample D. Dixon and Grubbs tests at levels 1 end 5 %, showed one laboratory outlier.
The outlier has been excluded from the calculation.
It is worth noting that the results obtained by FAME method were never considered as outliers by the tests applied.
Precision parameters
Tables 1 and 2 report the results of all the laboratories and the precision parameters calculated on an acceptable number (8) of labs but, unfortunately not deriving from the same analytical method.
Tables 3 and 4 report the results deriving only from TG method and the corresponding precision parameters. The acceptance of these parameters is subjected to the acceptance of the low number of laboratories (6).
Figures 2 and 3 show the trend of Sr and SR calculated on the 4 samples of the 2 data set described above.
Table 5 reports the Sr and SR values together with the corresponding pooled values and overall r and R parameters.
Finally the Critical Difference at 95 % of probability level has been calculated.
Table 1
Statistical Results of TG + FAME* methods
| Sample A | R1 | R2 | Mean | N. of labs retained after eliminating outliers | 8 | |
| RENNES | FR1 | 11,0 | 11,1 | 11,1 | N. of outliers | 1 |
| RIKILT | NL | 11,2 | 11,2 | 11,2 | Outliers | DК |
| ZPLA | DE* | 11,6 | 11,8 | 11,7 | Mean value | 11,3 |
| ADAS | GB | 11,4 | 11,2 | 11,3 | True value | 11,0 |
| CNEVA | FR2 | 11,4 | 11,4 | 11,4 | Repeatability standard deviation (Sr) | 0,09 |
| LODI | IT | 11,1 | 11,3 | 11,2 | Repeatability relative sd (RSDr%) | 0,80 |
| EELA | FI | 11,3 | 11,2 | 11,3 | Repeatability r (95 %) | 0,26 |
| ISPRA | UE* | 11,0 | 11,0 | 11,0 | Relative Repeatability r % | 2,24 |
| D.V.F.A. | DK | 13,3 | 11,8 | 12,6 | Reproducibility standard deviation (SR) | 0,23 |
| Reproducibility relative sd (RSDR%) | 2,04 | |||||
| Reproducibility R (95 %) | 0,84 | |||||
| Relative Reproducibility R % | 5,71 | |||||
| Sample B | R1 | R2 | Mean | N. of labs retained after eliminating outliers | 8 | |
| RENNES | FR1 | 12,7 | 12,8 | 12,8 | N, of outliers | 1 |
| RIKILT | NL | 13,5 | 13,3 | 13,4 | Outliers | DK |
| ZPLA | DE* | 14,0 | 13,8 | 13,9 | Mean value | 13,4 |
| ADAS | GB | 13,4 | 13,5 | 13,5 | True value | 13,5 |
| CNEVA | FR2 | 13,3 | 13,4 | 13,4 | Repeatability standard deviation (Sr) | 0,14 |
| LODI | IT | 13,9 | 13,5 | 13,7 | Repeatability relative sd (RSDr%) | 1,04 |
| EELA | FI | 13,4 | 13,2 | 13,3 | Repeatability r (95 %) | 0,40 |
| ISPRA | UE* | 13,2 | 13,3 | 13,3 | Relative Repeatability r % | 2,91 |
| D.V.F.A. | DK | 14,1 | 14,8 | 14,5 | Reproducibility standard deviation (SR) | 0,35 |
| Reproducibility relative sd (RSDR%) | 2,61 | |||||
| Reproducibility R (95 %) | 0,99 | |||||
| Relative reproducibility R % | 7,31 |
Table 2
Statistical Results of TG + FAME* methods
| Sample C | R1 | R2 | Mean | N. of labs retained after eliminating outliers | 8 | |
| RENNES | FR1 | 8,9 | 9,2 | 9,1 | N. of outliers | 1 |
| RIKILT | NL | 9,2 | 9,3 | 9,3 | Outliers | DK |
| ZPLA | DE* | 9,2 | 9,4 | 9,3 | Mean value | 9,3 |
| ADAS | GB | 9,5 | 9,3 | 9,4 | True value | 9,3 |
| CNEVA | FR2 | 9,4 | 9,4 | 9,4 | Repeatability standard deviation (Sr) | 0,14 |
| LODI | IT | 9,2 | 9,5 | 9,4 | Repeatability relative sd (RSDr%) | 1,50 |
| EELA | FI | 9,4 | 9,6 | 9,5 | Repeatability r (95 %) | 0,40 |
| ISPRA | UE* | 9,4 | 9,3 | 9,4 | Relative Repeatability r % | 4,20 |
| D.V.F.A. | DK | 10,7 | 10,9 | 10,8 | Reproducibility standard deviation (SR) | 0,17 |
| Reproducibility relative sd (RSDR%) | 1,82 | |||||
| Reproducibility R (95 %) | 0,47 | |||||
| Relative Reproducibility R % | 5,10 | |||||
| Sample D | R1 | R2 | Mean | N. of labs retained after eliminating outliers | 8 | |
| RENNES | R1 | 1,6 | 1,6 | 1,6 | N. of outliers | 1 |
| RIKILT | NL | 2,1 | 2,1 | 2,1 | Outliers | DK |
| ZPLA | DE* | 2,3 | 2,3 | 2,3 | Mean value | 2,1 |
| ADAS | GB | 2,1 | 2,2 | 2,2 | True value | 2,1 |
| CNEVA | FR2 | 2,1 | 2,1 | 2,1 | Repeatability standard deviation (Sr) | 0,08 |
| LODI | IT | 2,2 | 1,9 | 2,1 | Repeatability relative sd (RSDr%) | 3,81 |
| EELA | FI | 2,3 | 2,3 | 2,3 | Repeatability r (95 %) | 0,22 |
| ISPRA | UE* | 2,3 | 2,3 | 2,3 | Relative Repeatability r % | 10,67 |
| D.V.F.A. | DK | 3,4 | 2,9 | 3,2 | Reproducibility standard deviation (SR) | 0,24 |
| Reproducibility relative sd (RSDR%) | 11,43 | |||||
| Reproducibility R (95 %) | 0,67 | |||||
| Relative Reproducibility R % | 32,00 |
Table 3
Statistical Results of TG + FAME* methods
| Sample A | R1 | R2 | Mean | N. of labs retained after eliminating outliers | 6 | |
| RENNES | FR1 | 11,0 | 11,1 | 11,1 | N. of outliers | 1 |
| RIKILT | NL | 11,2 | 11,2 | 11,2 | Outliers | DК |
| ADAS | GB | 11,4 | 11,2 | 11,3 | Mean value | 11,2 |
| CNEVA | FR2 | 11,4 | 11,4 | 11,4 | True value | 11,0 |
| LODI | IT | 11,1 | 11,3 | 11,2 | Repeatability standard deviation (Sr) | 0,09 |
| EELA | FI | 11,3 | 11,2 | 11,3 | Repeatability relative sd (RSDr%) | 0,80 |
| D.V.F.A. | DK | 13,3 | 11,8 | 12,6 | Repeatability r (95 %) | 0,25 |
| Relative Repeatability r % | 2,24 | |||||
| Reproducibility standard deviation (SR) | 0,13 | |||||
| Reproducibility relative sd (RSDR%) | 1,16 | |||||
| Reproducibility R (95 %) | 0,36 | |||||
| Relative Reproducibility R % | 3,25 | |||||
| Sample B | R1 | R2 | Mean | N. of labs retained after eliminating outliers | 6 | |
| RENNES | FR1 | 12,7 | 12,8 | 12,8 | N. of outliers | 1 |
| RIKILT | NL | 13,5 | 13,3 | 13,4 | Outliers | DК |
| ADAS | GB | 13,4 | 13,5 | 13,5 | Mean value | 13,3 |
| CNEVA | FR2 | 13,3 | 13,4 | 13,4 | True value | 13,5 |
| LODI | IT | 13,9 | 13,5 | 13,7 | Repeatability standard deviation (Sr) | 0,15 |
| EELA | FI | 13,4 | 13,2 | 13,3 | Repeatability relative sd (RSDr%) | 1,13 |
| D.V.F.A. | DK | 14,1 | 14,8 | 14,5 | Repeatability r (95 %) | 0,42 |
| Relative Repeatability r % | 3,16 | |||||
| Reproducibility standard deviation (SR) | 0,33 | |||||
| Reproducibility relative sd (RSDR%) | 2,48 | |||||
| Reproducibility R (95 %) | 0,93 | |||||
| Relative Reproducibility R % | 6,94 |
Table 4
Statistical Results of TG method
| Sample C | R1 | R2 | Mean | N. of labs retained after eliminating outliers | 6 | |
| RENNES | FR1 | 8,9 | 9,2 | 9,1 | N. of outliers | 1 |
| RIKILT | NL | 9,2 | 9,3 | 9,3 | Outliers | DК |
| ADAS | GB | 9,5 | 9,3 | 9,4 | Mean value | 9,3 |
| CNEVA | FR2 | 9,4 | 9,4 | 9,4 | True value | 9,3 |
| LODI | IT | 9,2 | 9,5 | 9,4 | Repeatability standard deviation (Sr) | 0,15 |
| EELA | FI | 9,4 | 9,6 | 9,5 | Repeatability relative sd (RSDr%) | 1,61 |
| D.V.F.A. | DK | 10,7 | 10,9 | 10,8 | Repeatability r (95 %) | 0,42 |
| Relative Repeatability r % | 4,51 | |||||
| Reproducibility standard deviation (SR) | 0,19 | |||||
| Reproducibility relative sd (RSDR%) | 2,04 | |||||
| Reproducibility R (95 %) | 0,53 | |||||
| Relative Reproducibility R % | 5,71 | |||||
| Sample D | R1 | R2 | Mean | N, of labs retained after eliminating outliers | 6 | |
| RENNES | FR1 | 1,6 | 1,6 | 1,6 | N. of outliers | 1 |
| RIKILT | NL | 2,1 | 2,1 | 2,1 | Outliers | DK |
| Mean Value | 2,1 | |||||
| ADAS | GB | 2,1 | 2,2 | 2,2 | True value | 2,1 |
| CNEVA | FR2 | 2,1 | 2,1 | 2,1 | Repeatability standard deviation (Sr) | 0,09 |
| LODI | IT | 2,2 | 1,9 | 2,1 | Repeatability relative sd (RSDr%) | 4,29 |
| EELA | FI | 2,3 | 2,3 | 2,3 | Repeatability r (95 %) | 0,26 |
| D.V.F.A. | DK | 3,4 | 2,9 | 3,2 | Relative Repeatability r % | 12,01 |
| Reproducibility standard deviation (SR) | 0,25 | |||||
| Reproducibility relative sd (RSDR%] | 11,90 | |||||
| Reproducibility R (95 %) | 0,69 | |||||
| Relative Reproducibility R % | 33,32 |
Table 5
Repeatability and reproducibility (with FAME)
| CrD95 =0,40 Minimum purity stated for trienantoate = 95 % Minimum limit stated for trienantoate in butterfat = 11 kg/t Taking the Critical Difference for a 95 % probability level into consideration, the mean of the two results shall not be less than:
| ||||
| No of labs | Outlier | RepeatabilitySr (95 %) | ReproducibilitySR (95 %) | |
|---|---|---|---|---|
| Sample A | 8 | 1 | 0,09 | 0,23 |
| Sample Β | 8 | 1 | 0,14 | 0,35 |
| Sample C | 8 | 1 | 0,14 | 0,17 |
| Sample D | 8 | 1 | 0,08 | 0,24 |
| Pooled value | 0,116 | 0,256 | ||
| r | R | |||
| Pooled value* 2,8 | 0,324 | 0,716 | ||
Repeatability and reproducibility (without FAME)
| CrD95 = 0,36 Minimum purity stated for trienantoate = 95 % Minimum limit stated for trienantoate in butterfat = 11 kg/t Taking the Critical Difference for a 95 % probability level into consideration, the mean of the two results shall not be less than:
| ||||
| No of labs | Outlier | RepeatabilitySr (95 %) | ReproducibilitySR (95 %) | |
|---|---|---|---|---|
| Sample A | 6 | 1 | 0,09 | 0,13 |
| Sample B | 6 | 1 | 0,15 | 0,33 |
| Sample C | 6 | 1 | 0,15 | 0,19 |
| Sample D | 6 | 1 | 0,09 | 0,25 |
| Pooled value | 0,124 | 0,237 | ||
| r | R | |||
| Pooled value * 2,8 | 0,347 | 0,663 | ||
Figure 1(1)
Experimental results: Sample A
Experimental results: Sample B
Experimental results: Sample C
Experimental results: Sample D
= FAME method.